Page 60 - Fister jr., Iztok, and Andrej Brodnik (eds.). StuCoSReC. Proceedings of the 2015 2nd Student Computer Science Research Conference. Koper: University of Primorska Press, 2015
P. 60
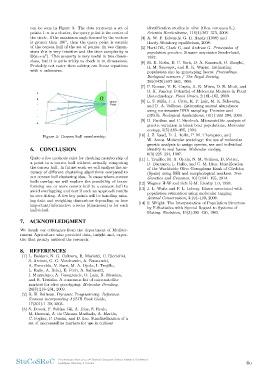
can be seen in Figure 3. The dots represent a set of identification studies in olive (Olea europaea L.).
points 1..n in a cluster, the query point is the center of Scientia Horticulturae, 116(4):367–373, 2008.
the circle. If the maximum angle formed by the vectors
is greater then 180◦, then the query point is outside [4] A. W. F. Edwards. G. H. Hardy (1908) and
of the convex hull of the set of points. In two dimen- Hardy-Weinberg equilibrium, 2008.
sions this is very intuitive and the time complexity is
O(m ∗ n2). This property is very useful in two dimen- [5] Hartl DL, Clark G, and Andrew G. Principales of
sions, but it is quite tricky to check in m dimensions. population genetics. Sinauer associates Sunderland,
Probably not easier then solving αm linear equations 1997.
with n unknowns.
[6] M. H. Kohn, E. C. York, D. A. Kamradt, G. Haught,
Figure 3: Convex hull membership R. M. Sauvajot, and R. K. Wayne. Estimating
population size by genotyping faeces. Proceedings.
6. CONCLUSION Biological sciences / The Royal Society,
266(1420):657–663, 1999.
Quite a few methods exist for checking membership of
a point in a convex hull without actually computing [7] P. Kumar, V. K. Gupta, A. K. Misra, D. R. Modi, and
the convex hull. In future work we will analyze the ac- B. K. Pandey. Potential of Molecular Markers in Plant
curacy of different clustering algorithms compared to Biotechnology. Plant Omics, 2:141–162, 2009.
our convex hull clustering idea. In cases where convex
hulls overlap we will explore the possibility of trans- [8] L. S. Mills, J. J. Citta, K. P. Lair, M. K. Schwartz,
forming one or more convex hull to a concave hull to and D. A. Tallmon. Estimating animal abundance
avoid overlapping and test if such an approach results using noninvasive DNA sampling: Promise and
in over-fitting. A few key points will be handling miss- pitfalls. Ecological Applications, 10(1):283–294, 2000.
ing data and weighting dimensions depending on how
important/informative a locus (dimension) is for each [9] D. Paetkau and C. Strobeck. Microsatellite analysis of
individual. genetic variation in black bear populations. Molecular
ecology, 3(5):489–495, 1994.
7. ACKNOWLEDGMENT
[10] J. Z. Reed, D. J. Tollit, P. M. Thompson, and
We thank our colleagues from the department of Mediter- W. Amos. Molecular scatology: the use of molecular
ranean Agriculture who provided data, insight and, exper- genetic analysis to assign species, sex and individual
tise that greatly assisted the research. identity to seal faeces. Molecular ecology,
6(3):225–234, 1997.
8. REFERENCES
[11] I. Trujillo, M. A. Ojeda, N. M. Urdiroz, D. Potter,
[1] L. Baldoni, N. G. Cultrera, R. Mariotti, C. Ricciolini, D. Barranco, L. Rallo, and C. M. Diez. Identification
S. Arcioni, G. G. Vendramin, A. Buonamici, of the Worldwide Olive Germplasm Bank of Co´rdoba
A. Porceddu, V. Sarri, M. A. Ojeda, I. Trujillo, (Spain) using SSR and morphological markers. Tree
L. Rallo, A. Belaj, E. Perri, A. Salimonti, Genetics and Genomes, 10(1):141–155, 2014.
I. Muzzalupo, A. Casagrande, O. Lain, R. Messina,
and R. Testolin. A consensus list of microsatellite [12] Wagner H-W and Sefc K-M. Identiy 1.0, 1999.
markers for olive genotyping. Molecular Breeding,
24(3):213–231, 2009. [13] J. L. Waits and P. L. Leberg. Biases associated with
population estimation using molecular tagging.
[2] R. E. Bellman. Dynamic Programming. Reference Animal Conservation, 3:191–199, 2000.
Reviews incorporating ASLIB Book Guide,
17(301):1–39, 2003. [14] S. Wright. The Interpretation of Population Structure
by F-Statistics with Special Regard to Systems of
[3] S. Doveri, F. Sabino Gil, A. D´ıaz, S. Reale, Mating. Evolution, 19(3):395–420, 1965.
M. Busconi, A. da Cˆamara Machado, A. Mart´ın,
C. Fogher, P. Donini, and D. Lee. Standardization of a
set of microsatellite markers for use in cultivar
StuCoSReC Proceedings of the 2015 2nd Student Computer Science Research Conference 60
Ljubljana, Slovenia, 6 October
points 1..n in a cluster, the query point is the center of Scientia Horticulturae, 116(4):367–373, 2008.
the circle. If the maximum angle formed by the vectors
is greater then 180◦, then the query point is outside [4] A. W. F. Edwards. G. H. Hardy (1908) and
of the convex hull of the set of points. In two dimen- Hardy-Weinberg equilibrium, 2008.
sions this is very intuitive and the time complexity is
O(m ∗ n2). This property is very useful in two dimen- [5] Hartl DL, Clark G, and Andrew G. Principales of
sions, but it is quite tricky to check in m dimensions. population genetics. Sinauer associates Sunderland,
Probably not easier then solving αm linear equations 1997.
with n unknowns.
[6] M. H. Kohn, E. C. York, D. A. Kamradt, G. Haught,
Figure 3: Convex hull membership R. M. Sauvajot, and R. K. Wayne. Estimating
population size by genotyping faeces. Proceedings.
6. CONCLUSION Biological sciences / The Royal Society,
266(1420):657–663, 1999.
Quite a few methods exist for checking membership of
a point in a convex hull without actually computing [7] P. Kumar, V. K. Gupta, A. K. Misra, D. R. Modi, and
the convex hull. In future work we will analyze the ac- B. K. Pandey. Potential of Molecular Markers in Plant
curacy of different clustering algorithms compared to Biotechnology. Plant Omics, 2:141–162, 2009.
our convex hull clustering idea. In cases where convex
hulls overlap we will explore the possibility of trans- [8] L. S. Mills, J. J. Citta, K. P. Lair, M. K. Schwartz,
forming one or more convex hull to a concave hull to and D. A. Tallmon. Estimating animal abundance
avoid overlapping and test if such an approach results using noninvasive DNA sampling: Promise and
in over-fitting. A few key points will be handling miss- pitfalls. Ecological Applications, 10(1):283–294, 2000.
ing data and weighting dimensions depending on how
important/informative a locus (dimension) is for each [9] D. Paetkau and C. Strobeck. Microsatellite analysis of
individual. genetic variation in black bear populations. Molecular
ecology, 3(5):489–495, 1994.
7. ACKNOWLEDGMENT
[10] J. Z. Reed, D. J. Tollit, P. M. Thompson, and
We thank our colleagues from the department of Mediter- W. Amos. Molecular scatology: the use of molecular
ranean Agriculture who provided data, insight and, exper- genetic analysis to assign species, sex and individual
tise that greatly assisted the research. identity to seal faeces. Molecular ecology,
6(3):225–234, 1997.
8. REFERENCES
[11] I. Trujillo, M. A. Ojeda, N. M. Urdiroz, D. Potter,
[1] L. Baldoni, N. G. Cultrera, R. Mariotti, C. Ricciolini, D. Barranco, L. Rallo, and C. M. Diez. Identification
S. Arcioni, G. G. Vendramin, A. Buonamici, of the Worldwide Olive Germplasm Bank of Co´rdoba
A. Porceddu, V. Sarri, M. A. Ojeda, I. Trujillo, (Spain) using SSR and morphological markers. Tree
L. Rallo, A. Belaj, E. Perri, A. Salimonti, Genetics and Genomes, 10(1):141–155, 2014.
I. Muzzalupo, A. Casagrande, O. Lain, R. Messina,
and R. Testolin. A consensus list of microsatellite [12] Wagner H-W and Sefc K-M. Identiy 1.0, 1999.
markers for olive genotyping. Molecular Breeding,
24(3):213–231, 2009. [13] J. L. Waits and P. L. Leberg. Biases associated with
population estimation using molecular tagging.
[2] R. E. Bellman. Dynamic Programming. Reference Animal Conservation, 3:191–199, 2000.
Reviews incorporating ASLIB Book Guide,
17(301):1–39, 2003. [14] S. Wright. The Interpretation of Population Structure
by F-Statistics with Special Regard to Systems of
[3] S. Doveri, F. Sabino Gil, A. D´ıaz, S. Reale, Mating. Evolution, 19(3):395–420, 1965.
M. Busconi, A. da Cˆamara Machado, A. Mart´ın,
C. Fogher, P. Donini, and D. Lee. Standardization of a
set of microsatellite markers for use in cultivar
StuCoSReC Proceedings of the 2015 2nd Student Computer Science Research Conference 60
Ljubljana, Slovenia, 6 October